科学计算
科学计算必备DataFrames
DataFrames基本操作
跟Python中的pandas的用法很像,相信用过Pandas的朋友上手应该无压力
DataFrame定义
新建一个DataFrame并增加4列内容
using DataFrames
df1 = DataFrame()
df1[:clo1] = Array([1.0,2.0,3.0])
df1[:clo2] = Array([4.0,5.0,6.0])
df1[:clo3] = Array([7.0,8.0,9.0])
df1[:ID] = Array(['a','b','c'])
show(df1)
>>3×4 DataFrame
│ Row │ clo1 │ clo2 │ clo3 │ ID │
│ │ Float64 │ Float64 │ Float64 │ Char │
├─────┼─────────┼─────────┼─────────┼──────┤
│ 1 │ 1.0 │ 4.0 │ 7.0 │ 'a' │
│ 2 │ 2.0 │ 5.0 │ 8.0 │ 'b' │
│ 3 │ 3.0 │ 6.0 │ 9.0 │ 'c' │
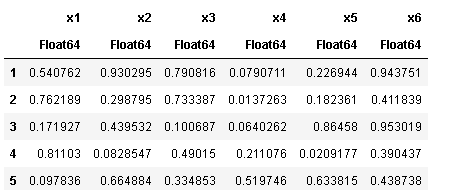
如果没有指定列名,则默认是x1,x2...
df2 = DataFrame(rand(5,6))
image
在DataFrame定义时直接指定内容
df3 = DataFrame([collect(1:3),collect(4:6)], [:A, :B])
>> A B
Int64 Int64
1 1 4
2 2 5
3 3 6
DataFrame合并
df2 = DataFrame()
df2[:clo1] = Array([11.0,12.0,13.0])
df2[:clo2] = Array([14.0,15.0,16.0])
df2[:clo3] = Array([17.0,18.0,19.0])
df2[:ID] = Array(['a','b','c'])
show(df2)
>>3×4 DataFrame
│ Row │ clo1 │ clo2 │ clo3 │ ID │
│ │ Float64 │ Float64 │ Float64 │ Char │
├─────┼─────────┼─────────┼─────────┼──────┤
│ 1 │ 11.0 │ 14.0 │ 17.0 │ 'a' │
│ 2 │ 12.0 │ 15.0 │ 18.0 │ 'b' │
│ 3 │ 13.0 │ 16.0 │ 19.0 │ 'c' │
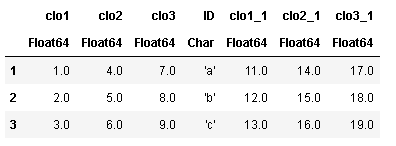
合并df1和df2
df3 = join(df1, df2, on=:ID)
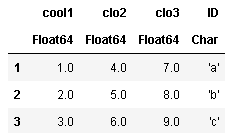
列重命名
rename!(df1, :clo1, :cool1)
索引
df = DataFrame(rand(5,6)) show(df)
5×6 DataFrame
│ Row │ x1 │ x2 │ x3 │ x4 │ x5 │ x6 │
│ │ Float64 │ Float64 │ Float64 │ Float64 │ Float64 │ Float64 │
├─────┼───────────┼──────────┼──────────┼──────────┼──────────┼───────────┤
│ 1 │ 0.678851 │ 0.514764 │ 0.829835 │ 0.996105 │ 0.427939 │ 0.163515 │
│ 2 │ 0.366487 │ 0.834486 │ 0.588859 │ 0.398066 │ 0.45738 │ 0.577453 │
│ 3 │ 0.0945619 │ 0.231903 │ 0.715664 │ 0.538696 │ 0.813534 │ 0.100683 │
│ 4 │ 0.102987 │ 0.269429 │ 0.292984 │ 0.719276 │ 0.756749 │ 0.705312 │
│ 5 │ 0.328692 │ 0.420872 │ 0.545552 │ 0.793303 │ 0.670403 │ 0.0619233 │
用索引取值,取24列
df[2:4]
>> x2 x3 x4
Float64 Float64 Float64
1 0.514764 0.829835 0.996105
2 0.834486 0.588859 0.398066
3 0.231903 0.715664 0.538696
4 0.269429 0.292984 0.719276
5 0.420872 0.545552 0.793303
用名称取值
df.x1
>>5-element Array{Float64,1}:
0.6788506862994854
0.366486647281848
0.09456191069734388
0.10298681965872736
0.32869200293154477
取24行,x1,x3列
df[2:4, [:x1, :x3]]
>> x1 x3
Float64 Float64
1 0.366487 0.588859
2 0.0945619 0.715664
3 0.102987 0.292984
for循环遍历
for row in eachrow(df)
println(row)
end
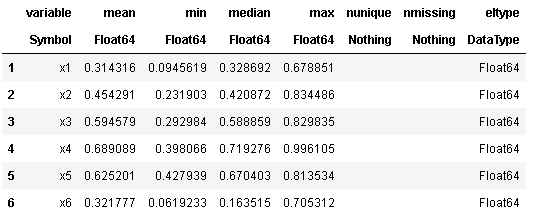
查看df的描述信息
describe(df)
排序
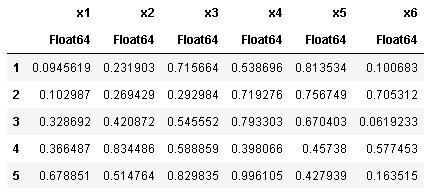
sort!(df, cols = [:x1])
DataFrame的csv文件操作
using CSV
df = DataFrame(rand(5,6))
show(df)
>>5×6 DataFrame
│ Row │ x1 │ x2 │ x3 │ x4 │ x5 │ x6 │
│ │ Float64 │ Float64 │ Float64 │ Float64 │ Float64 │ Float64 │
├─────┼──────────┼──────────┼───────────┼──────────┼───────────┼───────────┤
│ 1 │ 0.173904 │ 0.981004 │ 0.997001 │ 0.742527 │ 0.816245 │ 0.950086 │
│ 2 │ 0.946748 │ 0.661713 │ 0.0734147 │ 0.181932 │ 0.175604 │ 0.0550761 │
│ 3 │ 0.638618 │ 0.30026 │ 0.926336 │ 0.427942 │ 0.803738 │ 0.481783 │
│ 4 │ 0.156693 │ 0.943436 │ 0.0614211 │ 0.35279 │ 0.0692527 │ 0.417888 │
│ 5 │ 0.351843 │ 0.64204 │ 0.934611 │ 0.910804 │ 0.715309 │ 0.3677 │
写文件
CSV.write("df123.csv",df)
读文件
data = CSV.read("df123.csv")
DataFrame中的宏
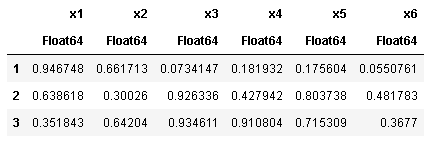
@where
using DataFramesMeta
@where(df, :x1 .> 0.2)
image
@with
@with(df, :x2 .+ 1)
>>5-element Array{Float64,1}:
1.9810036040861598
1.6617132284630372
1.3002596213068984
1.9434355174941438
1.6420398463078156
@select
@select(df, :x3)
>> x3
Float64
1 0.997001
2 0.0734147
3 0.926336
4 0.0614211
5 0.934611
@select(df, p = 2 * :x1, :x2)
>> p x2
Float64 Float64
1 0.347808 0.981004
2 1.8935 0.661713
3 1.27724 0.30026
4 0.313385 0.943436
5 0.703686 0.64204
RDatasets
RDatasets是Julia中的一个数据集,里面包含了很多可以学习和验证的数据,其中就包括iris数据集。
iris数据集介绍
在机器学习领域,有大量的公开数据集。iris就是其中非常重要的一个。
Iris Data Set(鸢尾属植物数据集)是一个历史很悠久的数据集,它首次出现在著名的英国统计学家和生物学家Ronald Fisher 1936年的论文《The use of multiple measurements in taxonomic problems》中,被用来介绍线性判别式分析。在这个数据集中,包括了三类不同的鸢尾属植物:Iris Setosa,Iris Versicolour,Iris Virginica。每类收集了50个样本,因此这个数据集一共包含了150个样本。
该数据集测量了所有150个样本的4个特征,分别是:
- sepal length(花萼长度)
- sepal width(花萼宽度)
- petal length(花瓣长度)
- petal width(花瓣宽度)
以上四个特征的单位都是厘米(cm)。
通常使用m表示样本量的大小,n表示每个样本所具有的特征数。
using RDatasets
iris = dataset("datasets", "iris")
features = convert(Array, iris[:, 1:4])
labels = convert(Array, iris[:, 5])
Gadfly绘图
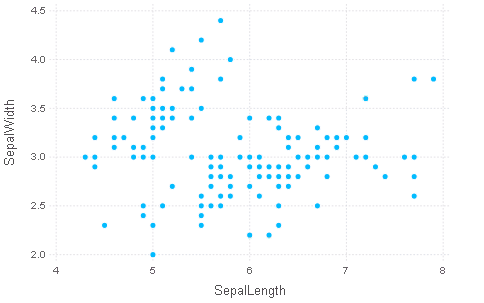
p = plot(iris, x=:SepalLength, y=:SepalWidth, Geom.point)
把图片输出到文件
img = SVG("iris_plot.SVG", 10cm, 8cm)
draw(img, p)
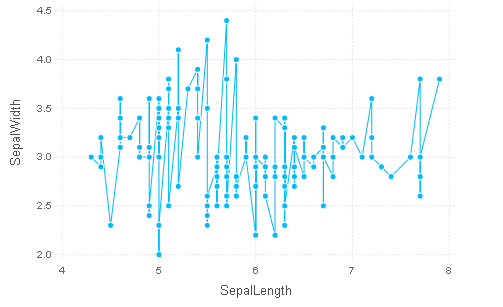
用线连接起来
p = plot(iris, x=:SepalLength, y=:SepalWidth, Geom.point, Geom.line)
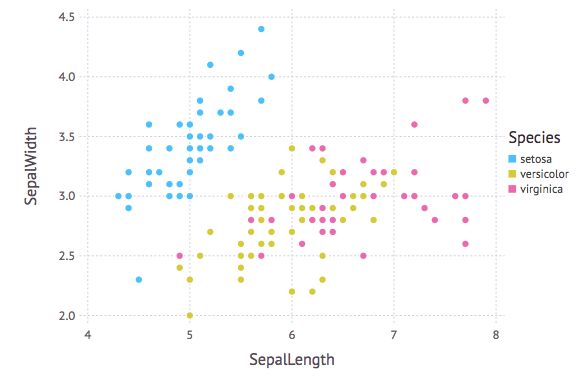
以Species作为颜色依据
p = plot(iris, x=:SepalLength, y=:SepalWidth, Geom.point, color=:Species)
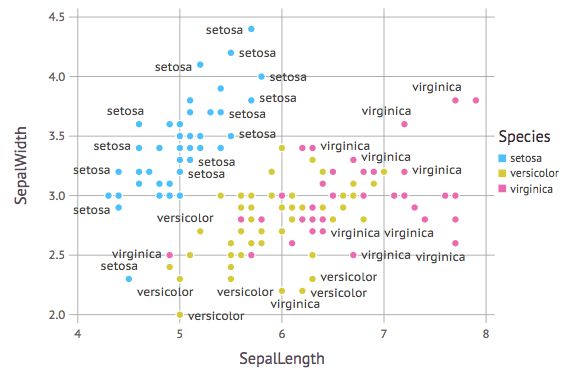
在图中加上lable
p = plot(iris, x=:SepalLength, y=:SepalWidth, label=:Species, color=:Species, Geom.point, Geom.label)
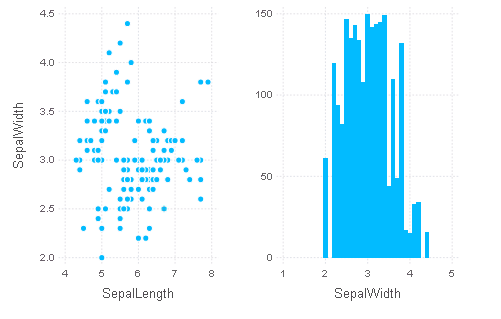
柱状图
fig1a = plot(iris, x="SepalLength", y="SepalWidth", Geom.point)
fig1b = plot(iris, x="SepalWidth", Geom.bar)
fig1 = hstack(fig1a, fig1b)
概率分布
using Distributions
生成均值20,标准差2.0的正态分布模型
n1 = Normal(20,2.0)params(n1)
>>(20,2.0)
fieldnames(typeof(n1))
>>(:μ, :σ)
Normal的类型是UnionAll
typeof(Normal)
>>UnionAll
生成满足n1模型的随机数
rand(n1,100)
二项分布
b = Binomial(20, 0.8)
rand(b, 1000)
概率密度
n = Normal()
pdf(n, 0.4)
>>0.36827014030332333
pdf(n, 0) #标准正态分布在x=0处的概率密度为0.3989...
>>0.3989422804014327
分布函数
cdf(n, 0.4)
>>0.6554217416103242
分位数
quantile.(n, [0.25, 0.5, 0.95])
>>3-element Array{Float64,1}:
-0.6744897501960818
0.0
1.6448536269514717
对于随机数rand(10),按Normal分布分析它的参数值
fit(Normal, rand(10))
>>Normal{Float64}(μ=0.41375573240875224, σ=0.25565343114046984)
StatsBase库:里面也包含了统计学的常用函数
using StatsBase
a = collect(1:6)
b = collect(4:9)
countne(a,b) #按顺序对比 1!=4 2!=5 ... 6!=9
>>6
a = [1,2,3,4,5]
b = [4,1,3,2,5]
counteq(a,b) # 按顺序对比 两个向量中相等元素的个数
>>2
L1dist(a,b) # abs(a[1]-b[1]) + ... + abs(a[n]-b[n])
>>6.0L2dist(a, b) # sqrt((a[1] - b[1])^2 + ... + (a[n] - b[n])^2)
meanad(a, b) # mean absolute deviation L1dist(a,b)/n
>>1.2
sample(a) # 从a中采样一次数据
sample(a, 3) # 从a中采样3次数据,返回1维Array
>>3-element Array{Int64,1}:
3
2
3
a1 = [1, 10, 20, 30]
a2 = [100, 200, 300]
sample!(a1, a2) # 从a1中,按照a2的类型取出length(a2)次数据
>>3-element Array{Int64,1}:
20
10
30
a1 = [1, 10, 20, 30]
a2 = [100.0, 200.0, 300.0]
sample!(a1, a2)
>>3-element Array{Float64,1}:
1.0
30.0
1.0
其他常用函数
x = [1.0, 2.0, 3.0, 4.0]
autocov(x)
autocov(x, [2,3])
autocor(x)
autocor(x, [2,3])
y = [3. , 4., 5., 6.]
crosscov(x, y)
crosscov(x, y, [2,3])
时间序列
using TimeSeries
新建Date变量
dates = collect(Date(2018, 1,1) :Day(1): Date(2018, 12, 31))
dates[1:20]
新建TimeArray
ta = TimeArray(dates, rand(length(dates),2))
>>365×2 TimeArray{Float64,2,Date,Array{Float64,2}} 2018-01-01 to 2018-12-31
│ │ A │ B │
├────────────┼────────┼────────┤
│ 2018-01-01 │ 0.7933 │ 0.4359 │
│ 2018-01-02 │ 0.6347 │ 0.7234 │
│ 2018-01-03 │ 0.2454 │ 0.6826 │
│ 2018-01-04 │ 0.767 │ 0.9007 │
│ 2018-01-05 │ 0.1884 │ 0.5429 │
│ 2018-01-06 │ 0.7809 │ 0.2292 │
│ 2018-01-07 │ 0.9329 │ 0.0098 │
│ 2018-01-08 │ 0.0032 │ 0.7566 │
│ 2018-01-09 │ 0.1355 │ 0.8986 │
│ 2018-01-10 │ 0.9274 │ 0.6022 │
│ 2018-01-11 │ 0.6483 │ 0.53 │
│ 2018-01-12 │ 0.8594 │ 0.0237 │
⋮
│ 2018-12-21 │ 0.7245 │ 0.5286 │
│ 2018-12-22 │ 0.9374 │ 0.8508 │
│ 2018-12-23 │ 0.0528 │ 0.1361 │
│ 2018-12-24 │ 0.1184 │ 0.6156 │
│ 2018-12-25 │ 0.3645 │ 0.7896 │
│ 2018-12-26 │ 0.023 │ 0.8259 │
│ 2018-12-27 │ 0.0942 │ 0.9326 │
│ 2018-12-28 │ 0.6124 │ 0.3102 │
│ 2018-12-29 │ 0.5602 │ 0.3305 │
│ 2018-12-30 │ 0.273 │ 0.7611 │
│ 2018-12-31 │ 0.7595 │ 0.1093 │
取出时间戳
timestamp(ta::TimeArray)
取值
values(ta::TimeArray)
取列名
colnames(ta::TimeArray)
机器学习
MLBase
using MLBase
机器学习基础库,不包含任何机器学习的算法,但为机器学习提供很多的必要工具,比如Cross validation等
先来看下MLBase中几个做简单数据处理的函数
repeach(1:3, 2)
>>6-element Array{Int64,1}:
1
1
2
2
3
3repeach(["a", "b", "c"], 2)
>>6-element Array{String,1}:
"a"
"a"
"b"
"b"
"c"
"c"
repeach(["a", "b", "c"], [1, 2, 3])
>>6-element Array{String,1}:
"a"
"b"
"b"
"c"
"c"
"c"
A = reshape(collect(1:6), 2, 3)
repeachcol(A, 2)
>>2×6 Array{Int64,2}:
1 1 3 3 5 5
2 2 4 4 6 6
repeachrow(A, 2)
>>4×3 Array{Int64,2}:
1 3 5
1 3 5
2 4 6
2 4 6
labels = labelmap(['a', 'b', 'b', 'c', 'c', 'c'])
>>LabelMap (with 3 labels):
[1] a
[2] b
[3] c
labelencode(labels, ['b', 'c', 'c', 'a'])
>>4-element Array{Int64,1}:
2
3
3
1
决策树
使用iris数据集
using RDatasets
iris = dataset("datasets", "iris")
features = convert(Array, iris[:, 1:4])
labels = convert(Array, iris[:, 5])
调用决策树
using DecisionTree
在该库中,包含了
- Decision Tree Classifier
- Random Forest Classifier
- Adaptive-Boosted Decision Stumps Classifier
- Regression Tree
- Regression Random Forest
我们这里只介绍分类模型,即前三种算法模型
model = build_tree(labels, features)
>>Decision Tree
Leaves: 9
Depth: 5
model = prune_tree(model, 0.9)
>>Decision Tree
Leaves: 8
Depth: 5
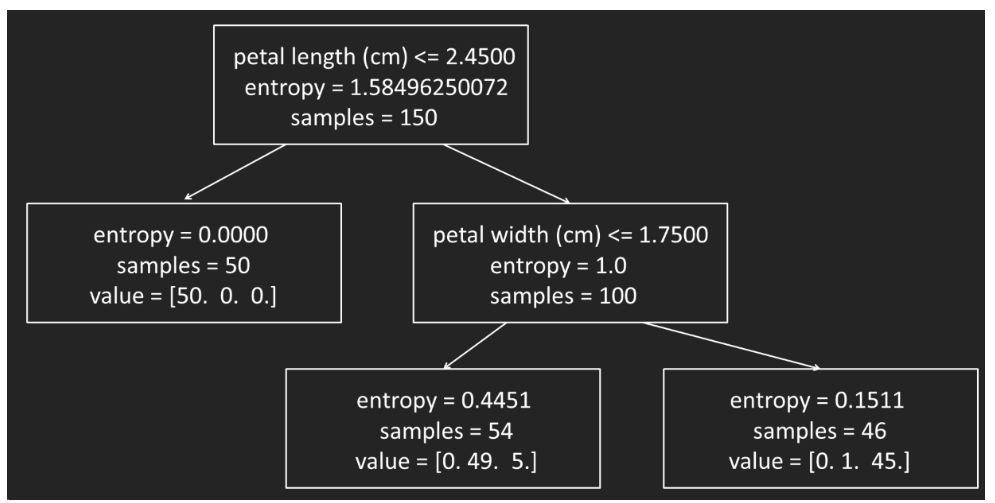
print_tree(model, 5)
>>Feature 3, Threshold 2.45
L-> setosa : 50/50
R-> Feature 4, Threshold 1.75
L-> Feature 3, Threshold 4.95
L-> versicolor : 47/48
R-> Feature 4, Threshold 1.55
L-> virginica : 3/3
R-> Feature 3, Threshold 5.449999999999999
L-> versicolor : 2/2
R-> virginica : 1/1
R-> Feature 3, Threshold 4.85
L-> Feature 2, Threshold 3.1
L-> virginica : 2/2
R-> versicolor : 1/1
R-> virginica : 43/43
print_tree(model, 3)
>>Feature 3, Threshold 2.45
L-> setosa : 50/50
R-> Feature 4, Threshold 1.75
L-> Feature 3, Threshold 4.95
L-> versicolor : 47/48
R->
R-> Feature 3, Threshold 4.85
L->
R-> virginica : 43/43
按照下面的方式
用决策树模型做判断
apply_tree(model, [5.9, 3.0, 5.1, 1.9])
>>"virginica"
apply_tree(model, [1.9, 2.0, 2.1, 1.9])
>>"setosa"
nfold cross validation for pruned tree
n_folds=3
accuracy = nfoldCV_tree(labels, features, n_folds, 0.9)
>>3×3 Array{Int64,2}:
24 0 0
0 13 0
0 0 13
3×3 Array{Int64,2}:
12 0 0
0 17 0
0 0 21
3×3 Array{Int64,2}:
14 0 0
0 20 0
0 1 15Fold 1
Classes: ["setosa", "versicolor", "virginica"]
Matrix:
Accuracy: 1.0
Kappa: 1.0Fold 2
Classes: ["setosa", "versicolor", "virginica"]
Matrix:
Accuracy: 1.0
Kappa: 1.0Fold 3
Classes: ["setosa", "versicolor", "virginica"]
Matrix:
Accuracy: 0.98
Kappa: 0.9695863746958637
Mean Accuracy: 0.9933333333333333
3-element Array{Float64,1}:
1.0
1.0
0.98
adaptive boosting
构造adaboost_stumps模型
model, coeffs = build_adaboost_stumps(labels, features, 10)
>>(Ensemble of Decision Trees
Trees: 10
Avg Leaves: 2.0
Avg Depth: 1.0, [0.346574, 0.255413, 0.264577, 0.335749, 0.288846, 0.258287, 0.356882, 0.298766, 0.242485, 0.349976])
应用模型判断
apply_adaboost_stumps(model, coeffs, [5.9, 3.0, 5.1, 1.9])
>>"virginica"
adaboost nfold cross-validation
# 3 folds, 8 iteration
accuracy = nfoldCV_stumps(labels, features, 3, 8)
随机森林
构造随机森林模型
# 2 random features, 10 trees, 0.5 portion of samples
model = build_forest(labels, features, 2, 10, 0.5)
>>Ensemble of Decision Trees
Trees: 10
Avg Leaves: 6.4
Avg Depth: 4.4
应用模型判断
apply_forest(model, [5.9, 3.0, 5.1, 1.9])
>>"virginica"
forest nfold cross-validation
accuracy = nfoldCV_forest(labels, features, 3, 2)
>>3×3 Array{Int64,2}:
12 0 0
0 15 0
0 1 22
3×3 Array{Int64,2}:
18 0 0
0 13 2
0 1 16
3×3 Array{Int64,2}:
20 0 0
0 20 0
0 0 10Fold 1
Classes: ["setosa", "versicolor", "virginica"]
Matrix:
Accuracy: 0.98
Kappa: 0.9689440993788819Fold 2
Classes: ["setosa", "versicolor", "virginica"]
Matrix:
Accuracy: 0.94
Kappa: 0.9096385542168673Fold 3
Classes: ["setosa", "versicolor", "virginica"]
Matrix:
Accuracy: 1.0
Kappa: 1.0
Mean Accuracy: 0.9733333333333333
3-element Array{Float64,1}:
0.98
0.94
1.0
MultivariateStats
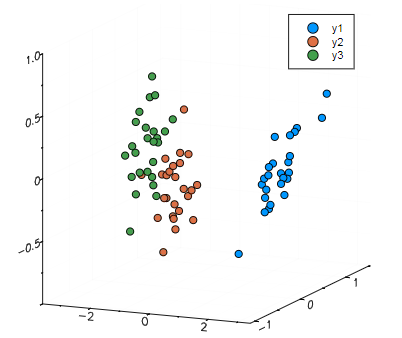
PCA(Principal Component Analysis)是一种常用的数据分析方法。PCA通过线性变换将原始数据变换为一组各维度线性无关的表示,可用于提取数据的主要特征分量,常用于高维数据的降维。
using MultivariateStats, RDatasetsload iris dataset
iris = dataset("datasets", "iris")
split half to training set
Xtr = convert(Array,(iris[1:2:end,1:4]))'
Xtr_labels = convert(Array,(iris[1:2:end,5]))split other half to testing set
Xte = convert(Array,(iris[2:2:end,1:4]))'
Xte_labels = convert(Array,(iris[2:2:end,5]))suppose Xtr and Xte are training and testing data matrix,
with each observation in a column
train a PCA model, allowing up to 3 dimensions
M = fit(PCA, Xtr; maxoutdim=3)
apply PCA model to testing set
Yte = transform(M, Xte)
reconstruct testing observations (approximately)
Xr = reconstruct(M, Yte)
group results by testing set labels for color coding
setosa = Yte[:,Xte_labels.=="setosa"]
versicolor = Yte[:,Xte_labels.=="versicolor"]
virginica = Yte[:,Xte_labels.=="virginica"]
size(Xte)
>>(4, 75)
size(Yte)
>>(3, 75)
把降维后的数据画出来
using Plots
p = scatter(setosa[1,:],setosa[2,:],setosa[3,:],marker=:circle,linewidth=0)
scatter!(versicolor[1,:],versicolor[2,:],versicolor[3,:],marker=:circle,linewidth=0)
scatter!(virginica[1,:],virginica[2,:],virginica[3,:],marker=:circle,linewidth=0)
SVM
using LIBSVM
using RDatasets
iris = dataset("datasets", "iris")
features = convert(Array, iris[:, 1:4])
labels = convert(Array, iris[:, 5])
features_train, features_test = features[1:2:end, :], features[2:2:end, :]
labels_train, lables_test = labels[1:2:end], labels[2:2:end]
model = svmtrain(features_train', labels_train)
(predicted_labels, decision_values) = svmpredict(model, features_test')
检查预测结果
using Statistics
mean(predicted_labels .== lables_test) * 1.0
predicted_labels .== lables_test
>>75-element BitArray{1}:
true
true
true
true
true
true
true
true
true
true
true
true
true
⋮
true
true
true
true
true
true
true
true
true
true
true
true
线性回归
using GLM, GLMNet, DataFrames
lm模型
data = DataFrame(X=[1,2,3], Y=[2,4,7])
ols = lm(@formula(Y ~ X), data)
>>StatsModels.DataFrameRegressionModel{LinearModel{LmResp{Array{Float64,1}},DensePredChol{Float64,LinearAlgebra.Cholesky{Float64,Array{Float64,2}}}},Array{Float64,2}}Formula: Y ~ 1 + X
Coefficients:
Estimate Std.Error t value Pr(>|t|)
(Intercept) -0.666667 0.62361 -1.06904 0.4788
X 2.5 0.288675 8.66025 0.0732
线性回归模型标准差
stderror(ols)
>>2-element Array{Float64,1}:
0.6236095644623245
0.2886751345948133
应用模型
newX = DataFrame(X=[2,3,4]);
GLM.predict(ols, newX)
>>3-element Array{Union{Missing, Float64},1}:
4.333333333333332
6.833333333333335
9.33333333333334
data = DataFrame(X=[1,2,3], Y=[2,4,6])
ols = lm(@formula(Y ~ X), data)
newX = DataFrame(X=[2,3,4])
GLM.predict(ols, newX)
>>3-element Array{Union{Missing, Float64},1}:
4.0
6.0
8.0
glm模型
data = DataFrame(X=[0,1,2,3,4], Y=[0.1296,0.0864,0.0576,0.0384,0.0256])
probit = glm(@formula(Y ~ X), data, Binomial(), ProbitLink())
>>StatsModels.DataFrameRegressionModel{GeneralizedLinearModel{GlmResp{Array{Float64,1},Binomial{Float64},ProbitLink},DensePredChol{Float64,LinearAlgebra.Cholesky{Float64,Array{Float64,2}}}},Array{Float64,2}}Formula: Y ~ 1 + X
Coefficients:
Estimate Std.Error z value Pr(>|z|)
(Intercept) -1.14184 1.32732 -0.860259 0.3896
X -0.208282 0.659347 -0.315891 0.7521
newX = DataFrame(X=[1,2,3,4])
GLM.predict(probit, newX)
>>4-element Array{Union{Missing, Float64},1}:
0.08848879660655808
0.05956904945792997
0.03864063839150974
0.024136051585243873
线性回归模型处理iris数据集
data = DataFrame();
data[:x] = iris[1:50, :SepalWidth]
data[:y] = iris[1:50, :SepalLength]
model = lm(@formula(y ~ x), data)
>>StatsModels.DataFrameRegressionModel{LinearModel{LmResp{Array{Float64,1}},DensePredChol{Float64,LinearAlgebra.Cholesky{Float64,Array{Float64,2}}}},Array{Float64,2}}Formula: y ~ 1 + x
Coefficients:
Estimate Std.Error t value Pr(>|t|)
(Intercept) 2.639 0.310014 8.51251 <1e-10
x 0.69049 0.0898989 7.68074 <1e-9
把LM拟合后的曲线跟原始的曲线做对比
using Plots
plot(GLM.predict(model))
plot!(data[:y])
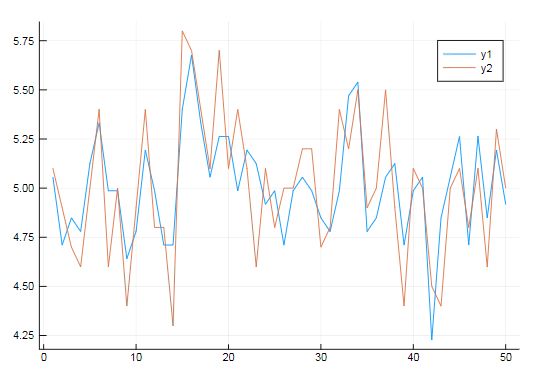
用GLM处理iris数据集
model = glm(@formula(y~x), data, Normal(), IdentityLink())
>>StatsModels.DataFrameRegressionModel{GeneralizedLinearModel{GlmResp{Array{Float64,1},Normal{Float64},IdentityLink},DensePredChol{Float64,LinearAlgebra.Cholesky{Float64,Array{Float64,2}}}},Array{Float64,2}}Formula: y ~ 1 + x
Coefficients:
Estimate Std.Error z value Pr(>|z|)
(Intercept) 2.639 0.310014 8.51251 <1e-16
x 0.69049 0.0898989 7.68074 <1e-13
把GLM拟合后的曲线跟原始的曲线做对比
using Plots
plot(GLM.predict(model))
plot!(data[:y])
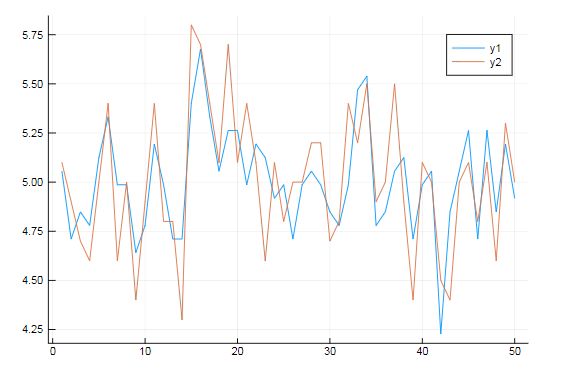
# Estimates for coefficents
coef(model)Standard errors of coefficents
stderror(model)
Covariance of coefficents
vcov(model)
RSS
deviance(model)
K-means
using Clustering
using RDatasets
iris = dataset("datasets", "iris")
features = convert(Array, iris[:, 1:4])
labels = convert(Array, iris[:, 5]);
# choose 3 random starting points
initseeds(:rand, convert(Matrix,features'), 3)
>>3-element Array{Int64,1}:
48
80
114
result = kmeans(features, 3)
>>KmeansResult{Float64}([5.1 2.45 0.2; 4.9 2.2 0.2; … ; 6.2 4.4 2.3; 5.9 4.05 1.8], [1, 2, 2, 3], [3.63798e-12, 166.128, 166.127, 0.0], [1, 2, 1], [1.0, 2.0, 1.0], 332.2550000000019, 2, true)
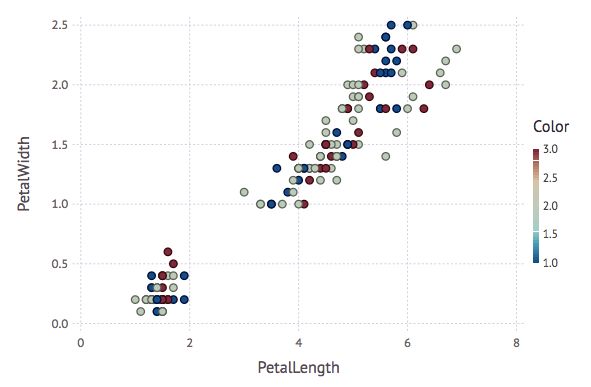
using Gadfly
plot(iris,x="PetalLength",y="PetalWidth",color=result.assignments, Geom.point)